Overview
Our lab is interested in the function, regulation, and targeting of enzymes that control protein post-translational modifications (PTM). After the human genome is sequenced, a major challenge to understand biology and human diseases is to understand the function of all proteins. Proteins are not always expressed, and even if expressed, their activities are not always on. Their activities are often regulated by certain stresses and signals, which is crucial for all cell signaling events. Thus, to fully understand the function of a protein, it is also crucial to understand how it is regulated. We, therefore, focus on understanding the function and regulation of PTM enzymes and use the understanding to develop small molecule inhibitors for these enzymes as potential therapeutics for treating human diseases, including cancer, autoimmune diseases, and neurodegenerative diseases.
NAD-consuming enzymes: Sirtuins and PARPs
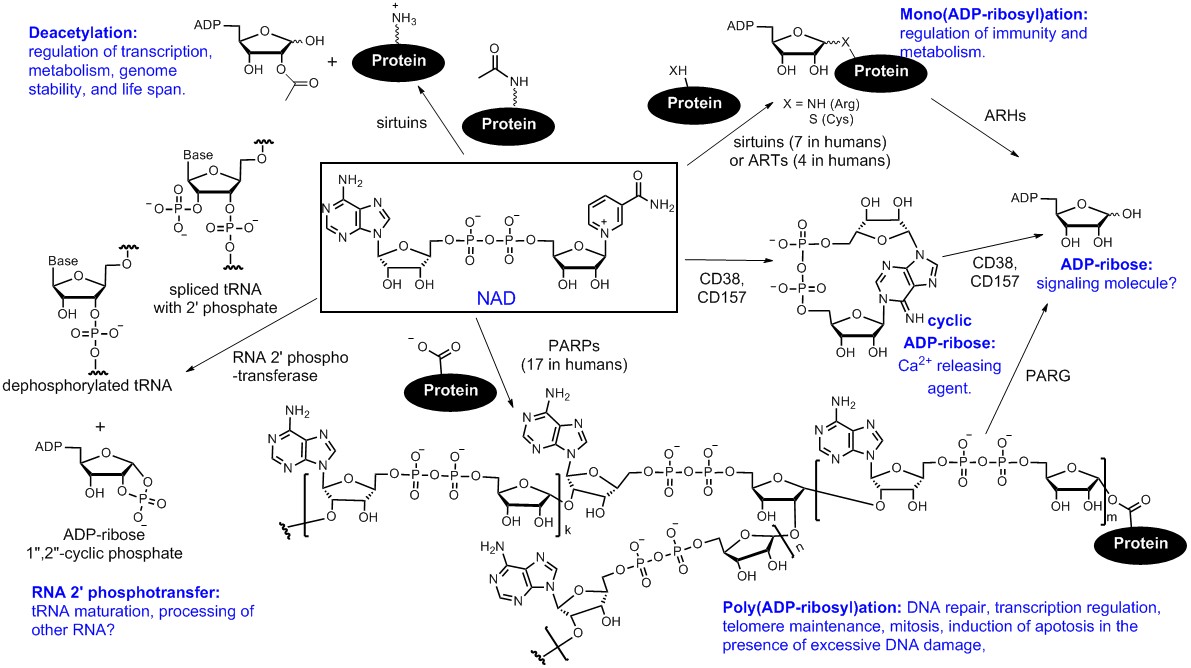
Among the PTM enzymes that we study, we are particularly interested in NAD-consuming enzymes. These enzymes stand out as they not only have diverse and important biological functions but also display very interesting chemistry. Figure 1 summarizes some of the known NAD-consuming enzymatic reactions, including three that modify proteins: NAD-dependent deacetylation, mono(ADP-ribosyl)ation, and poly(ADP-ribosyl)ation.
Figure 1. Known cellular enzymatic reactions that consume NAD. These reactions include: (1) Poly(ADP-ribosyl)ation, catalyzed by poly(ADP-ribose) polymerases (PARPs); (2) mono(ADP-ribosyl)ation, catalyzed by both sirtuins, certain PARPs, and ADP-ribosyltransferases (ARTs); (3) NAD-dependent deacetylation, catalyzed by sirtuins; (4) removal of 2′-phosphate from tRNA splicing intermediates, catalyzed by RNA 2′-phosphotransferases; (5) formation of cyclic ADP-ribose, catalyzed by both CD38 and CD157 in mammals. Some of the biological functions of these NAD-consuming reactions are also indicated in the figure.
Sirtuins and HDACs. The Sir2 (silencing information regulator 2) family of enzymes, or sirtuins, were originally known as NAD-dependent protein lysine deacetylases (Figure 2). They are present in all domains of life and have been shown to be important in regulating numerous biological pathways, including genome stability, metabolism, and longevity. Mammals have seven sirtuin enzymes, Sirt1-7. They are considered promising targets for treating several human diseases, including cancer and neurodegeneration. Among the seven mammalian sirtuins, only Sirt1-3 have efficient deacetylase activity. Sirt4-7 have very weak and sometimes undetectable deacetylase activity. We demonstrated that Sirt5 can remove succinyl and malonyl groups while Sirt6 can remove myristoyl and palmitoyl groups very efficiently (Figure 2). We demonstrated that protein lysine succinylation, malonylation, and long-chain fatty acylation are common protein posttranslational modifications (PTMs) that were previously unknown or under-recognized. We are continuing to understand the functions of sirtuins by discovering new substrates and their regulatory mechanisms. Furthermore, we are also utilizing information about the enzymatic activity of sirtuins to develop small molecule inhibitors for therapeutic applications.
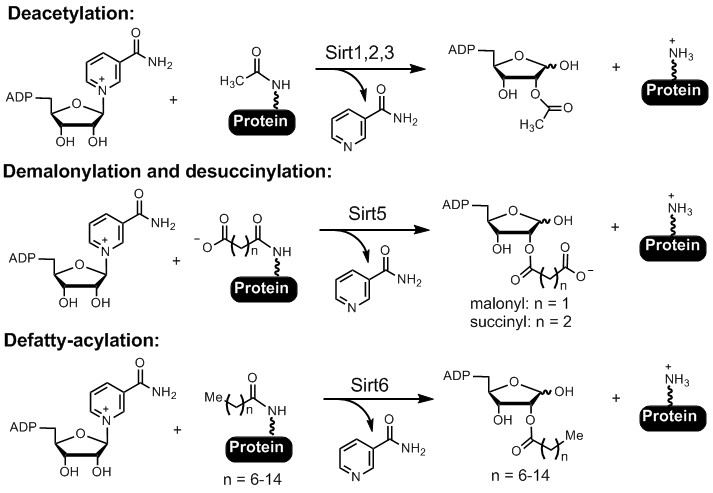 Figure 2. Sirtuin-catalyzed deacylation reactions.
Figure 2. Sirtuin-catalyzed deacylation reactions.
Another class of enzymes closely related to sirtuins are the zinc-dependent histone deacetylases or HDACs. There are 11 HDACs in humans and many have very weak or no detectable deacetylase activity (HDAC4, 5, 7, 8, 9, 11). We are also interested in studying the function of these HDACs.
Representative publications on sirtuins and HDACs:
- J. Cao et al, ,“HDAC11 regulates type I interferon signaling through defatty-acylation of SHMT2”, Proc Natl Acad Sci USA. 116:5487-5492, 2019
- N.A. Spiegelman et al., “SIRT2 and Lysine Fatty Acylation Regulate the Activity of RalB and Cell Migration”, ACS Chem Biol. 14:2014-2023, 2019.
- J.Y. Hong et al., “A Glycoconjugated SIRT2 Inhibitor with Aqueous Solubility Allows Structure-Based Design of SIRT2 Inhibitors”, ACS Chem Biol. 14:1802-1810, 2019.
- H. Jing et al., SIRT2 and lysine fatty acylation regulate the transforming activity of K-Ras4a. Elife. pii: e32436, 2017. doi: 10.7554/eLife.32436
- X. Zhang et al., “SIRT6 regulates Ras-related protein R-Ras2 by lysine defatty-acylation.” Elife. 6. pii: e25158, 2017.
- X. Zhang et al., “Identifying the functional contribution of the defatty-acylase activity of SIRT6”, Chem. Biol. 12:614-20, 2016.
- P. Aramsangtienchai et al., “HDAC8 Catalyzes the Hydrolysis of Long Chain Fatty Acyl Lysine.” ACS Chem. Biol. 11:2685-2692, 2016.
- H. Jing et al., “A SIRT2-specific inhibitor promotes c-Myc oncoprotein degradation and inhibits tumor growth”, Cancer Cell, 29: 297–310, 2016.
- H. Sadhukhan et al., “ Metabolomics-assisted Proteomics Identifies Succinylation and SIRT5 as Important Regulators of Heart Function”, Proc. Natl. Acad. Sci. USA, 113:4320-5, 2016.
- H. Jiang et al., “Sirt6 regulates TNFa secretion via hydrolysis of long chain fatty acyl lysine”, Nature, 496, 110-113 (2013).
- B. He, J. Du, H. Lin, “Thiosuccinyl Peptides as Sirt5-Specific Inhibitors”, J. Am. Chem. Soc., 134, 1922-1925 (2012).
- A. Y. Zhu, Y. Zhou, S. Khan, K. W. Deitsch, Q. Hao, H. Lin, “Plasmodium falciparum Sir2A preferentially hydrolyzes medium and long chain fatty acyl lysine”, ACS Chem. Biol., 7, 155-159 (2012).
- J. Du, Y. Zhou, X. Su, J. Yu, Khan S., H. Jiang, J. Kim, J. Woo, J.H. Kim, B.H. Choi, B. He, W. Chen, S. Zhang, R.A. Cerione, J. Auwerx, Q. Hao, H. Lin, “Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase”, Science, 334, 806-809 (2011).
PARPs. PARPs catalyze either poly-ADP-ribosylation or mono-ADP-ribosylation of various substrate proteins. There are 17 PARPs in humans,For most other PARPs, their biological functions are still unknown. Many PARP inhibitors are in clinical trial for treating cancers, especially triple-negative breast cancers. To better realize the potential of PARP inhibitors as therapeutics, it is important to understand the biological functions of various PARPs. Our earlier work focused on identifying the substrate proteins of PARPs. We developed clickable NAD analogs to identify PARP substrate proteins (Figure 3). The clickable NAD analog has an alkyne functional group, which allows the conjugation, via click chemistry, of different tags, such as fluorescent tags for in-gel visualization and affinity tags for purification. Furthermore, we demonstrate that by identifying the substrate proteins of PARP1, novel insights into its biological functions can be obtained.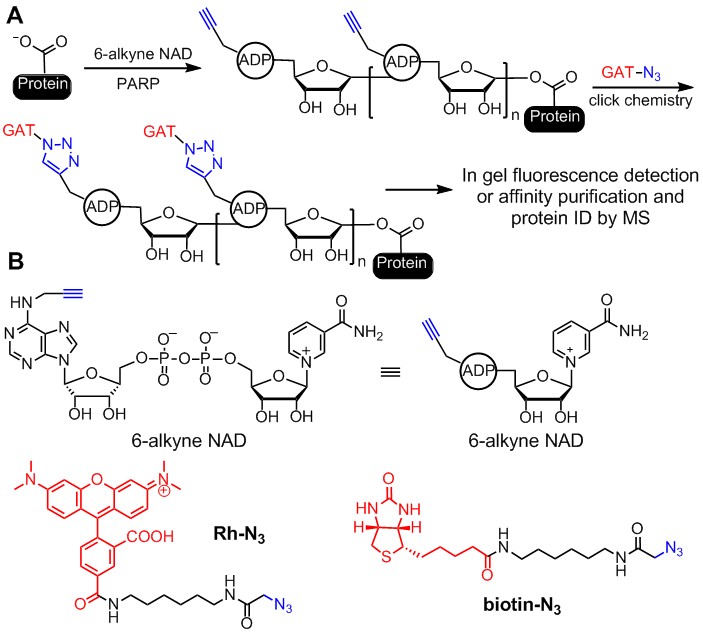
Figure 3. Labeling the substrate proteins of PARPs using clickable NAD analogs. (A) Labeling of PARylated proteins with 6-alkyne NAD. An affinity tag can be added using click chemistry after the substrate protein is labeled. The labeled protein can then be affinity purified, separated on 1D/2D protein gel, and then the sequence identified by MS. (B) Structures of compounds used in labeling reactions.
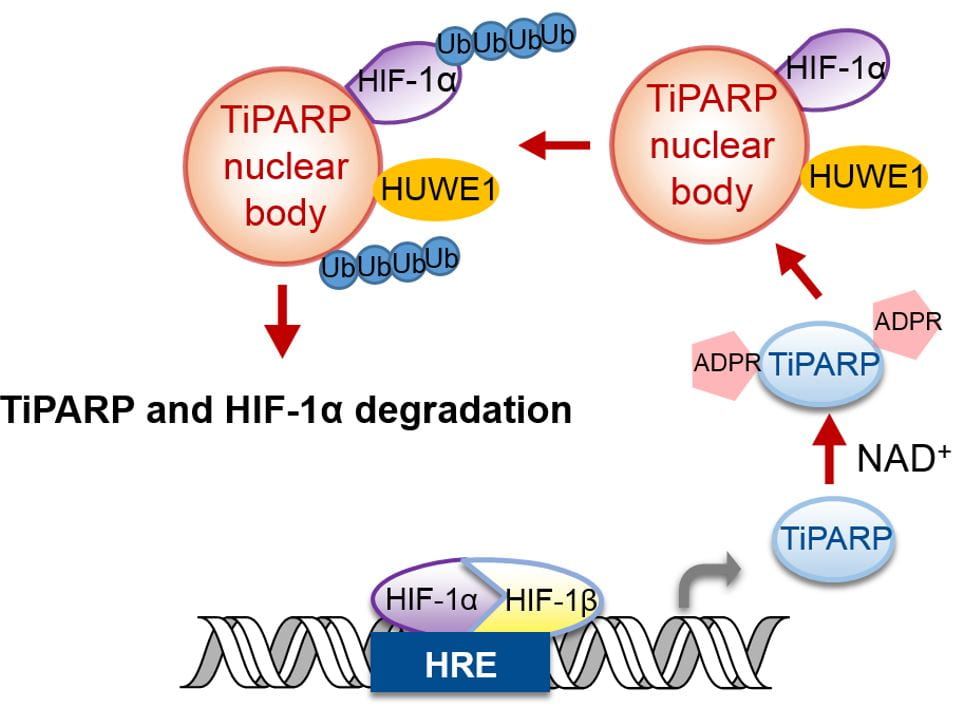
Recently, we started to study the function of PARPs by understanding their regulation. Our work with TiPARP (PARP7) showed that it is transcriptionally up-regulated by HIF-1. The increased TiPARP expression serves to ADP-ribosylate HIF-1α and promote HIF-1α degradation, forming a negative feedback loop to turn off HIF-1 transcriptional activity (Figure 4). The most interesting part of this story is the mechanism via which TiPARP promotes the degradation of HIF-1α. We found that TiPARP, via its ADP-ribosylation activity, forms phase condensates (nuclear bodies) and recruits E3 ubiquitin ligases, thus leading to the ubiquitination and degradation of HIF-1α. We are currently investigating the detailed mechanism via which ADP-ribosylation promotes phase condensation.
Figure 4. Model for the regulation of HIF-1 by TiPARP-mediated ADP-ribosylation, phase condensation, and ubiquitination.
Representative publications on PARPs:
- Zhang L, Cao J, Dong L, Lin H. TiPARP forms nuclear condensates to degrade HIF-1α and suppress tumorigenesis. Proc Natl Acad Sci U S A. 117:13447-13456, 2020
- H. Jiang, J. Congleton, Q. Liu, P. Merchant, F. Malavasi, H.C. Lee, Q. Hao, A. Yen, H. Lin, “Mechanism-based fluorescent labeling of human CD38”, J. Am. Chem. Soc., 131, 1658-1659 (2009).
ZDHHC and NMT long-chain acyltransferases
Our interest in sirtuins and HDACs led us to study the function of protein lysine long-chain acylation. In this process, we got interested in the long-chain acylation of other protein residues, such as cysteine and N-terminal glycine. Two families of enzymes are well-known to catalyze these long-chain acylations, ZDHHC family for cysteine palmitoylation and NMT for N-terminal glycine myristoylation. Our recent work surprisingly showed that NMT could also catalyze N-terminal lysine side chain myrisotylation on ARF6, This myristoylation can be removed by SIRT2 and this myristoylation-demyristoylation cycle is important for the activity of ARF6. We are currently working to discover other substrate proteins that are lysine myristoylated by NMT.
Similarly, we found another acylation-deacylation cycle for STAT3. STAT3 can be cysteine palmitoylated by ZDHHC7, which targets STAT3 to the plasma membrane, where it can be phosphorylated by JAK2. The phosphorylated STAT3 then is depalmitoylated by APT2, allowing phosphorylated STAT3 to go to the nucleus to turn on transcription of genes required for Th17 development. Thus, the palmitoylation-depalmitoylation cycle of STAT3 is crucial for its function and immune activation. We have shown that pharmacologically or genetically disrupting this cycle can effectively inhibit Th17 cell differentiation and suppress inflammatory bowel diseases. We are currently studying various ZDHHCs by discovering their substrate proteins and understand how cysteine palmitoylation regulates their functions.
Representative publications on ZDHHC and NMT:
- M. Zhang et al., A STAT3 palmitoylation cycle promotes Th17 differentiation and colitis, Nature, 586:434-439, 2020.
- T. Kosciuk et al., “NMT1 and NMT2 are Lysine Myristoyltransferases Regulating the ARF6 GTPase Cycle” Nat. Commun. 11:1067, 2020
Dipthamide: biosynthesis and biological function
Diphtheria was once a deadly disease causing many deaths before modern vaccination was available. The disease is caused by Corynebacterium diphtheriae, a bacterium that secrets diphtheria toxin. Diphtheria toxin catalyzes the ADP-ribosylation of a unique posttranslationally-modified His residue, termed diphthamide, in eukaryotic and archaeal translation elongation factor 2 (EF2). EF2 is a GTPase that catalyzes the translocation of the peptidyl-tRNA and mRNA from the ribosome A site to the P site, and therefore is essential for protein biosynthesis. The biological function of diphthamide is not understood yet, although it has been shown that knock out dph1 or dph2 is lethal in mouse, and dph1 heterozygote mouse are prone to tumor formation. The biosynthesis of the diphthamide residue has been a long time puzzle. There are at least seven genes (Dph1, Dph2, Dph3, Dph4, Dph5, Dph6, and Dph7) required for the biosynthesis. Our lab contributed to the discovery of Dph6 and Dph7.
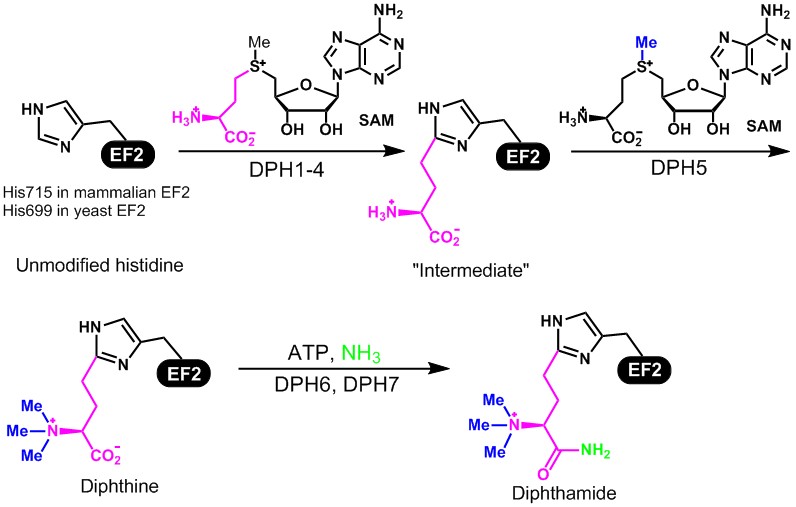
The proposed biosynthesis pathway is shown in Figure 5. Our goal is to use in vitro biochemistry to figure out the molecular functions of the proteins Dph1-7 in the biosynthesis of diphthamide, and study the effects of diphthamide formation on the function of EF2 in protein synthesis. The first step of the biosynthesis is of particular interest because of the uncommon C-C bond formation reaction, the uncommon use of SAM, and the requirement of multiple proteins (Dph1-4). Our evidences suggest that the enzyme catalyzing this step contains [4Fe-4S] cluster and uses an unusual radical reaction mechanism. Our work has led to a much better understanding of how this unusual [4Fe-4S] radical enzyme works. Our current focus is on understanding the biological function of this modification. Specifically, we are interested to find out the translation of what proteins are affected by diphthamide and what the cellular phenotypes are when diphthamide biosynthesis is disrupted.
Figure 5. The proposed biosynthesis pathway for diphthamide and its ADP-ribosylation by diphtheria toxin.
Representative publications on Diphthamide:
- M. Dong et al., Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis, Science, 359:1247-1250, 2018
- M. Dong et al., Substrate-Dependent Cleavage Site Selection by Unconventional Radical SAM Enzymes in Diphthamide Biosynthesis. J Am Chem Soc. 139:5680-5683, 2017.
- Z. Lin et al., “Cbr1 is a Dph3 reductase required for the tRNA wobble uridine modification.” Nat Chem Biol. 12:995-997, 2016.
- M. Dong et al., “Organometallic Complex Formed by an Unconventional Radical S-Adenosylmethionine Enzyme.” J Am Chem Soc. 138:9755-9758, 2016.
- Z. Lin et al., “Dph7 catalyzes a previously unknown demethylation step in diphthamide biosynthesis”. J. Am. Chem. Soc. 136, 6179-6182, 2014
- M. Dong et al., “Dph3 is an electron donor for dph1-dph2 in the first step of eukaryotic diphthamide biosynthesis”, J. Am. Chem. 136, 1754, 2014.
- X. Su, Z. Lin, W. Chen, H. Jiang, S. Zhang, and H. Lin, “A chemogenomic approach identified yeast YLR143W as diphthamide synthetase”, Proc. Natl. Acad. Sci. USA, 109, 19983-19987 (2012).
- X. Su, W. Chen, W. Lee, H. Jiang, S. Zhang, H. Lin, “YBR246W Is Required for the Third Step of Diphthamide Biosynthesis”, J. Am. Chem. Soc.,134, 773-776 (2012)
- Y. Zhang, X. Zhu, A. Torelli, M. Lee, B. Dzikovski, R. M. Koralewski, E. Wang, J. Freed, C. Krebs, S. E. Ealick, H. Lin, “Diphthamide biosynthesis requires an Fe-S enzyme-generated organic radical”, Nature, 465, 891-896 (2010).